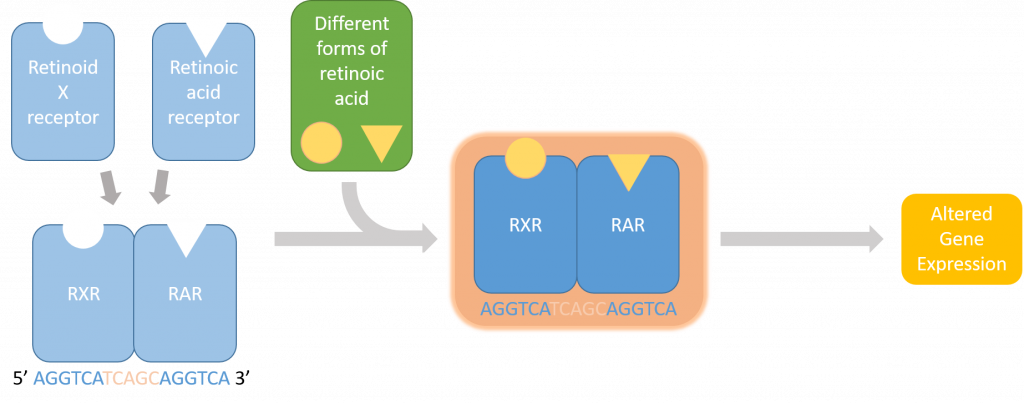
Hi everyone! My name is Jiarong Li and I’m a senior majoring in math and computer science at Reed. During this summer, in order to offer reference information for computational researchers to choose a suitable cloud computing platform, I worked with Anna and Trina to compare mainly four cloud computing platforms: Google Cloud, Amazon Web Service (AWS), Extreme Science and Engineering Discovery Environment (XSEDE), and the Institute for Bioinformatics and Evolutionary Studies from UIdaho (iBEST CRC).
By running three cloud computing projects related to different fields, I analyzed the real application of these platforms in terms of the execution time, computing capacity, storage, customizability, compatibility, and convenience. I designed a scoring rubric and tabulated and summarized the testing results with various plots.
Projects and Testing Results
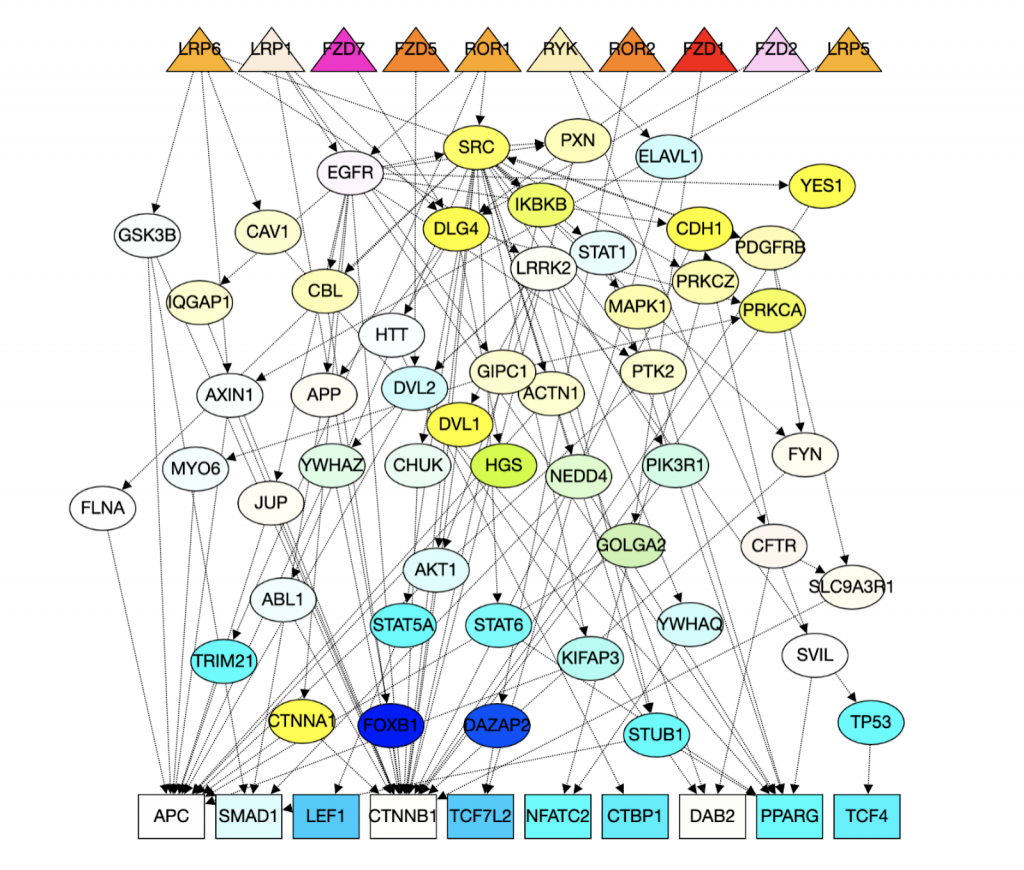
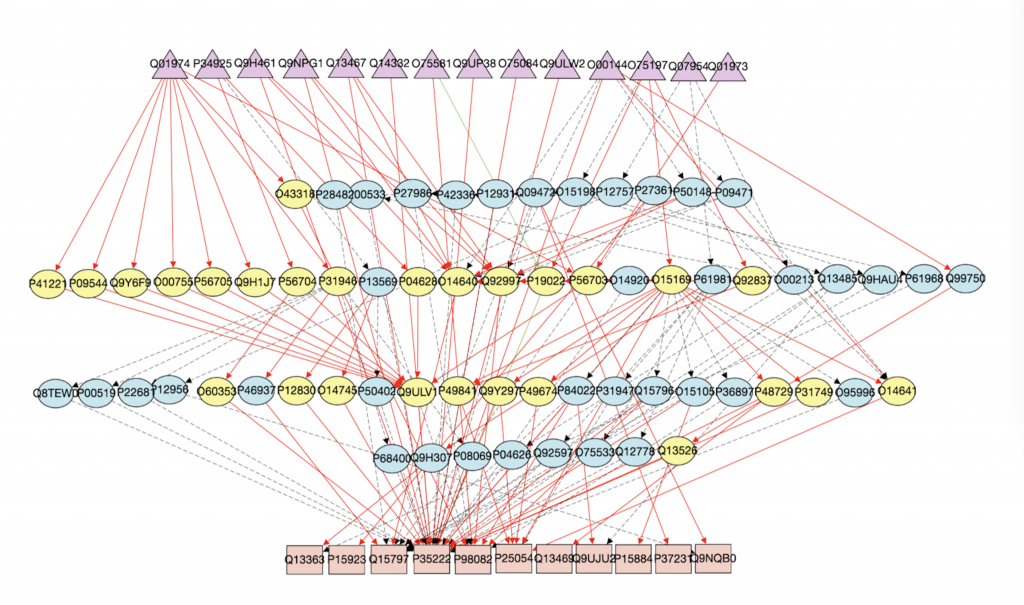
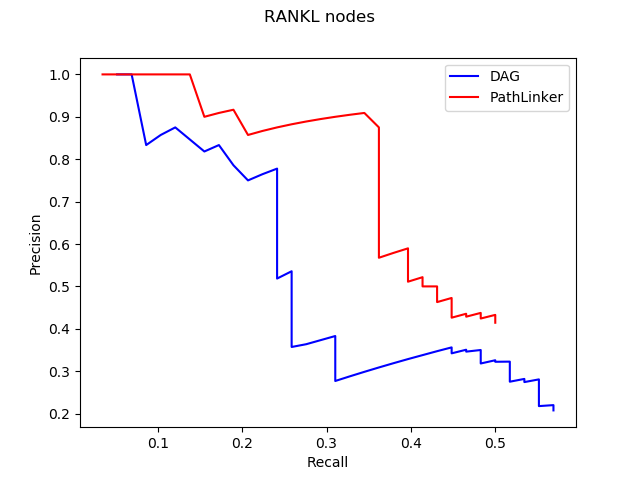
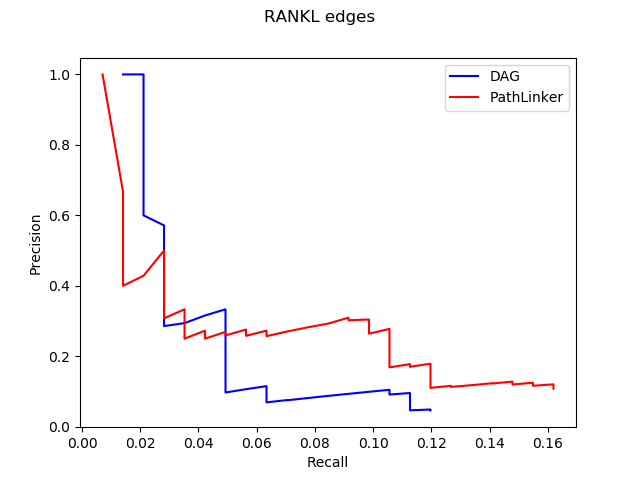
1. Computational Biology: Augmenting signaling pathway reconstructions (PRAUG) [1]
By running PRAUG to reconstruct signaling pathways and recoding the runtime, I focused on testing the computing ability of the same type of virtual machines from different platforms.
Testing Result
The trend shown in the graph above tells us that virtual machines with more vCPUs take less execution time. Since PRAUG code was originally set to run on a 4-core machine, without setting up multithread, the trend of the decreasing execution time cannot be seen when the number of vCPUs getting above 6.
In the graphs above, I mainly compared the performance of virtual machines from Google Cloud and AWS since they have comparable customizability. With the same settings of virtual machines, we can see that AWS outperforms Google Cloud.
XSEDE and iBEST have less customizability and option of virtual machines. I chose several log-in nodes to run the program and their testing results are:
14 CPUs & 128 GB: 13m33.907s (XSEDE)
16 CPUs & 3 TB: 23m5.172s (XSEDE)
40 CPUs & 192 GB: 31m28.872s (iBEST)
2. Large Image Files Subsampling and Transferring: Tabular data files for 3D still images
By running multithreaded R code to randomly generate a specified number of 3D image subsamples with a certain number of pixels, this project mainly focuses on testing virtual machines with multiple vCPUs as well as the capability and convenience of data transferring and storing of different platforms.
Testing Result
The graph above shows the difference of execution time when we input the same variables and run the single-threaded code and multi-threaded code respectively on a virtual machine with 8 cores and 64GB memory.
In this project, by running multithreaded code, we can clearly see the influence of the number of vCPUs to the execution time.
When I set the number of subsamples to 20 and pixels/sample to 100, the runtime of this R code on VMs from Google Cloud and AWS are shown above. Still, AWS outperforms Google Cloud in some extent. Moreover, I tested more resources provided by XSEDE and iBEST in this project and Jetstream from XSEDE has really good performance.
In addition to multithreaded computing, I also compared the storage and data transferring of these four platforms. I personally like the s3 bucket from AWS the best since it is very convenient to access the files stored in s3 through other platforms and the security settings of private files have great customizability. Data transferring can be easily achieved by Globus among these four platforms and other applications.
3. Machine Learning: Running a machine learning experiment in a specific Docker Image
This project mainly focuses on testing GPU virtual machines and their compatibility with other applications.
Testing Result
The graph above shows the comparison of runtime when training the ML model on VMs with NVIDIA Tesla T4 GPU. We can see that VMs from AWS run faster than those from Google Cloud.
I also tried to adopt VMs with different types of GPUs and got results shown in the graph above. For more information about NVIDIA Tesla.
I didn’t run this project on GPU nodes from XSEDE or iBEST since I have no permission to set up docker on these VMs. Generally, for all four platforms, users need to request access to GPU VMs and will get either permission or rejection depending on specific situation.
Summary
Based on all the testing results and the user experience I got during this process, I evaluated the four platforms according to the scoring rubric designed at the beginning.
Thanks to Anna, Tobias, Kelly, and Mark for providing cloud computing projects tested in this research.
References
[1] Rubel, Tobias, and Anna Ritz. “Augmenting Signaling Pathway Reconstructions.” BioRxiv. January 01, 2020. Accessed August 29, 2020. https://www.biorxiv.org/content/10.1101/2020.06.16.155853v1.full.