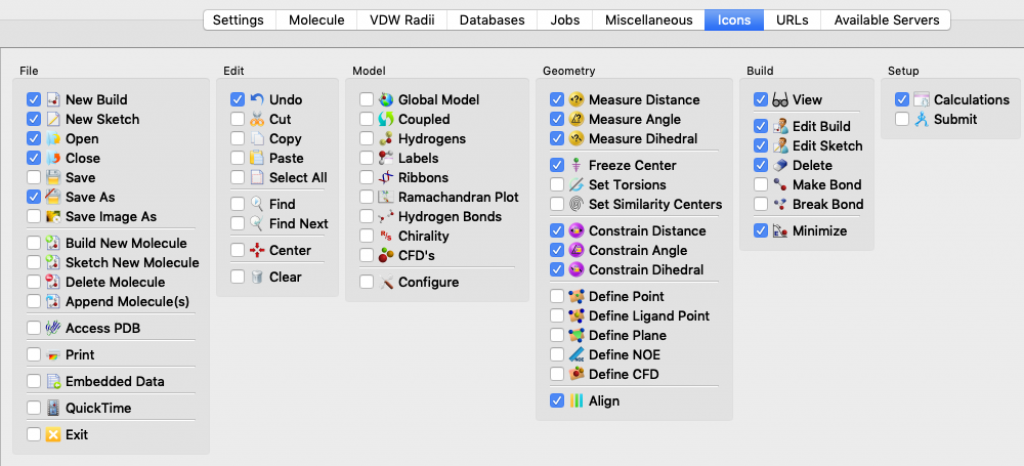
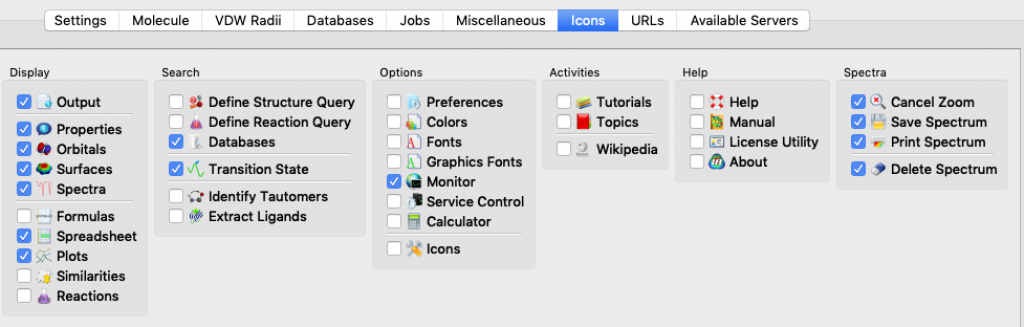
I mentioned this topic briefly during a modeling session, but it went by fast and you may have missed the key instructions. The idea here is that Spartan lets you choose which icons to display in your Spartan window so that you can tailor things to match your needs and work more efficiently.
Because every icon corresponds to a menu item, you might think to yourself, “Why clutter up my window? I’ll just rely on the menus.” That would be a mistake. Not only do the icons save you mouse clicks, they also provide visual reminders about the tools that are available and work flow.
Here’s how you select icons for an Apple (Mac) computer. Choose Options > Preferences. When the Preferences window opens, choose the ICONS tab (it turns blue). The icons are listed by menu. Start with the File menu, click on the icons you want, and then continue working your way, menu by menu, all the way over to the Spectra menu, 12 menus in all. (Notes: 1) the Spectra menu doesn’t actually appear in the Spartan window, 2) my window doesn’t display all 12 menus at once so I had to use the slider at the bottom of the window to bring more menus into view.) When you’re done, click OK.
These thumbnail images show the icons that I currently rely on (click on the thumbnail for a more legible image):

Recent Comments