Though there has been great progress with understanding actin and microtubule dynamics, and their relationship to cell motility, the scientific community is still working to try to better define these interactions, and their associated proteins, that allow this movement to happen. Many neurodevelopmental disorders and degenerative diseases are linked to genes associated with the cytoskeleton (Prokop et. al., 2013), making impaired motility of neurons a possible cause. Therefore, further information on growth and motility dynamics could help lead potential treatments for these disorders.
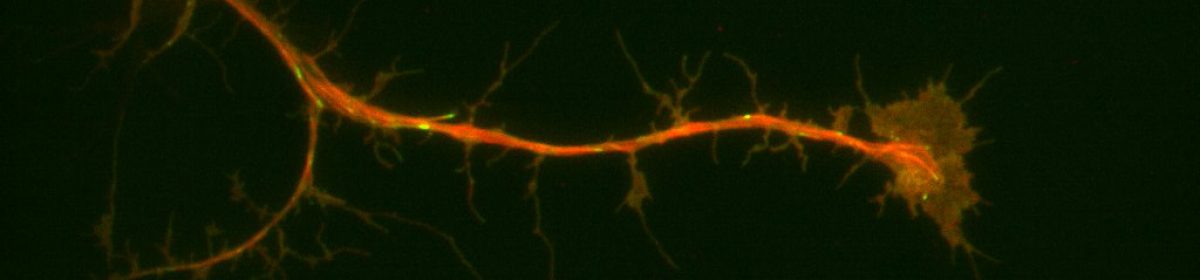
Neurons are understood to develop an axon, their outwardly-signaling appendage, by the elongation of a neurite via microtubule recruitment. The growth cone at the tip of this developing axon is actin-rich, and forms focal adhesions with the substrate to facilitate movement and network development (Prokop et. al., 2013), making the growth cone the leading edge in neurons. Interestingly, there have been some indications that these growth cones may move at different rates on some substrates than others. Because of the substrate-dependency lead, it may be possible to utilize different substrates to characterize some of the proteins present in the extracellular matrix that contribute to efficient movement of neurons. For my thesis project I’m attempting to look at growth cone dynamics on two substrates: extracellular matrix (ECM) and concanavalin A (ConA, a plant lectin often used to plate cells) using neurons extracted from Drosophila 3rd instar larvae.
Drosophila lend themselves as an excellent model for examining the role of proteins in neuronal movement. Not only are the proteins affecting actin and microtubule dynamics well-conserved, making the findings generalizable across species, but the genetics are simplified in drosophila as well, with little redundancy in the genome. This attribute makes it so that a single knockout is capable of almost completely eliminating the function of a protein, whereas in other animals, such as mice, the knockout of multiple genes may be required to silence a protein due to redundancy (Prokop et. al., 2013). Additionally, existing knowledge about their genome makes it so that we can easily and time-efficiently make genetically altered strains for experimentation. This makes knockdown studies easy and efficacious, and desirable crosses quick to generate. Their neurons have been found to grow well in cultures, even forming networks and displaying similar electrical properties to in-vivo functioning neurons when in proper media (Küppers-Munther et. al., 2004). This allows easy visualization and analysis of neuronal movement, further contributing to their experimental value, which is why drosophila were chosen by our lab for these studies.
In a typical experiment, I extract 6-10 brains from the drosophila larvae and apply liberase to break apart the cell-cell contacts. These neurons are then plated in rich cell media on either an ECM- or a ConA-coated glass slide in a dish and allowed to attach to the bottom of the slide. After the neurons have attached, I visualize their movement by taking videos under a microscope. Additional visualization of proteins can be achieved with staining.
Sources:
Küppers-Munther, B., Letzkus, J. J., Lüer, K., Technau, G., Schmidt, H., & Prokop, A. (2004). A new culturing strategy optimises Drosophila primary cell cultures for structural and functional analyses. Dev Biol, 269(2), 459-478.
Prokop, A., Beaven, R., Qu, Y., & Sánchez-Soriano, N. (2013). Using fly genetics to dissect the cytoskeletal machinery of neurons during axonal growth and maintenance. J Cell Sci, 126(Pt 11), 2331-2341.