My name is Tommy Yoon (he/him) and I’m a class of 2024 BMB major. This summer I had the opportunity to continue the work of Max and Henry in finding manganese homeostasis proteins in a collaboration with Shivani Ahuja. This summer, instead of working on a multi-species network for manganese and zinc homeostasis, we decided to focus the question on manganese homeostasis in Bacillus subtilis, a gram-positive model bacteria. We used motif finding, transcriptomic analysis, and a protein-protein interaction network to find other proteins involved in manganese homeostasis, and to find links to other systems which sense manganese or other ions.
Manganese Homeostasis
B. subtilis maintains manganese homeostasis with a manganese sensor MntR, which regulates the transcription of uptake proteins MntABCD and MntH, and efflux proteins MneP and MneS.
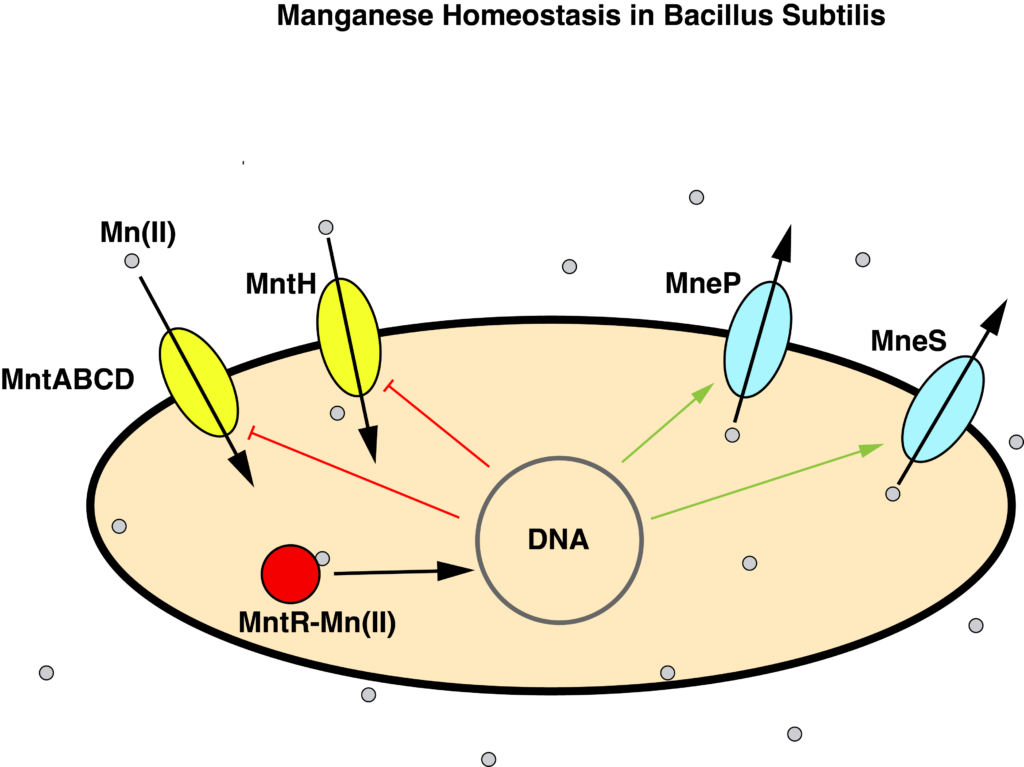
Motif Finding
We started by looking for the binding motif of MntR in promoter regions of the B. subtilis genome. The idea was that genes with the binding motif would probably be regulated by MntR. We implemented several iterations of a motif finder, including one that looked for clusters of motifs, since MntR often binds in clusters. However, since the information content of the motif is so low, multiple occurrences of the motif showed up before pretty much every gene. It was clear that there are other factors influencing MntR binding, so we moved on to other methods.
Transcriptomics
John Helmann from Cornell graciously supplied some gene expression data from wild-type and mutant mntR samples in both low (50 nM) and high (2.5 µm) concentrations of manganese. By analyzing this data, we can find genes that respond to the absence of mntR and manganese concentration. We looked for genes which responded to a combination of those two factors. Surprisingly, we found a lot of iron uptake genes in this analysis upregulated by a combination of high Mn(II) and mntR mutation. We already knew that high Mn(II) leads to increased expression of iron genes, but this analysis may implicate mntR in the regulation of iron homeostasis.
Network Analysis
We used the STRING protein-protein interaction network to find genes that may be involved in manganese homeostasis. To do this, we used two algorithms: random walk with restarts (RWR) and Omics Integrator. The principle behind these algorithms is that proteins involved in the same process tend to interact with each other more often. If we find out which proteins are interacting with known manganese homeostasis proteins, we may find some candidate proteins that contribute to manganese homeostasis.
RWR works as follows: start with a source node and walk randomly to connected nodes, keeping track of which nodes are visited. At each step, restart with some probability at another source node. The nodes that get visited the most are strongly connected to the source nodes. Omics Integrator has a similar goal to find nodes that connect your source nodes. Since the known manganese proteins are not very well-connected, we used the differentially expressed genes as our set of source nodes for both of these algorithms. This analysis found a number of central proteins. One surprising find was a magnesium transport protein YfjQ. It is known that magnesium levels are increased in mntR mutants, and so the implication of the magnesium transporter YfjQ in manganese response systems is an interesting result!
Below is a sample Omics Integrator output using downregulated genes as source nodes. Red indicates source nodes and node size indicates how many times the node was found over 900 replicates with different algorithm parameters and source node sets.
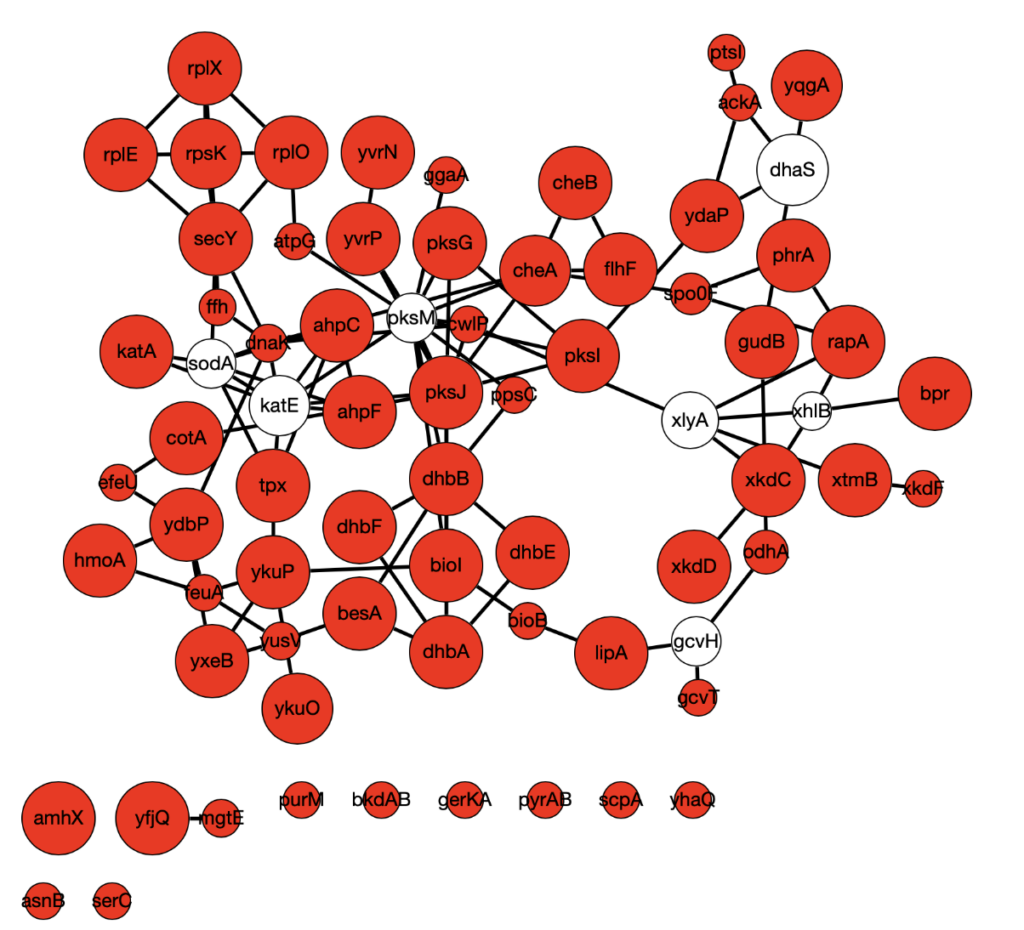
There is more computational work to be done here to narrow down on a list of candidates, and more data is always being generated. The door is also open at this point for wet lab work to investigate some of the genes identified in our screen. This is a fun project—I learned a lot about a lot of different things and I’m immensely grateful for the opportunity to work on this project. Big thanks to Anna and the rest of the lab for a great summer!
