Modelling MntABC and ZnuABC Transporter Regulation with Protein Interaction Data from the STRING Database
Hello reader! My name is Max Bennett, I’m a Senior Neuroscience major at Reed College and I’ve spent the summer of 2022 in Anna Ritz’s CompBio lab implementing a protein interactome model to learn more about bacterial metal ion transporters. This work was a collaborative effort with Biochemistry Professor Shivani Ahuja and Junior Biochemistry major Henry Jacques.
Metal Ion Transporters MntABC and ZnuABC
Many transition metals are critical cofactors for biological enzymatic reactions in animals like you and me, but also in bacteria. As cofactors, the properties of each metal ion are very unique, and one cannot be substituted for another, but they are all 2+ metal ions of similar size and weight, so proteins designed to transport or sequester these valuable resources often have affinity for multiple metal ions. For pathogenic bacteria this is a liability that makes them especially vulnerable to their host manipulating the concentrations of these ions up or down to induce toxicity or starvation. The human immune system primarily manipulates Zn (II), critical in its own right, but also a competitor for Mn (II), which is a cofactor for reducing reactive oxygen species (ROS); too much Zn (II) will outcompete Mn (II) and the cell will succumb to oxidative stress. The ATP-Binding Cassette (ABC) transporters MntABC and ZnuABC are at the heart of this system and have both been proven critical for pathogenic bacteria to successfully withstand an immune response and cause infection. Understanding this metal ion system could let us eventually target and interfere with it, posing a very promising new class of antibiotic drugs.
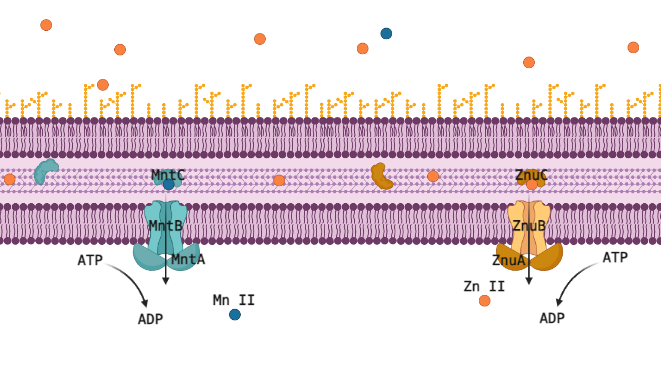
The Zn (II) and Mn (II) transport systems are incredibly sensitive, suggesting a fast-acting regulatory system that can rapidly respond to changes in the environmental and internal concentrations of these ions and to the needs of the cell. Professor Shivani Ahuja’s structural biochemistry lab is well equipped to capture this regulatory interaction red handed, but the involved proteins must be identified and purified to do that. We wouldn’t know which proteins to purify though, the present literature just doesn’t have strong candidates for post translational regulation of metal ion ABC transporters. This is where computational methods offer the chance to use disparate data from multiple species to piece together regulatory mechanisms for MntABC and ZnuABC.
The Protein Interactome
Protein interactomes are network models (also called graphs) of the proteins in an organism and the interactions between them. Obviously, the word interactions is vague, and usefully so even: it can make just as much sense to show genetic regulation interactions, co-expression, cytogenetic closeness, sequence similarity, and other kinds of biologically relevant relationships besides what I’ll call “physical interactions,” but at the heart of protein interactomes are those physical interactions. A physical interaction refers to 2 proteins associating together as a complex or in an allosteric interaction; the gold standard for proving this kind of interaction is the protein pull-down assay. Protein interaction data in the STRING database is the cornerstone of our analysis. The first thing we set out to do was to recreate the single species protein interactome models in Python using data from STRING and to make a very human-legible and well integrated interface for the interactome on Graphspace. Below is an example of a Graphspace interactome alongside the protein information and linked databases available for every node.
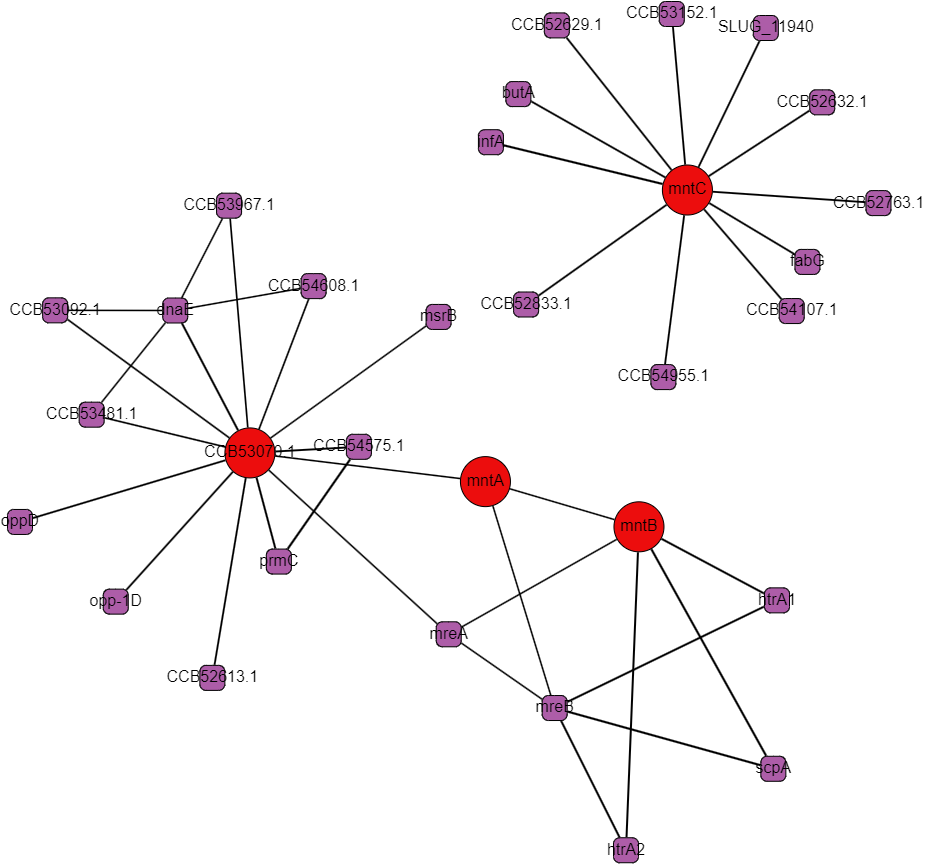
STRING’s interaction data is broad in scope but it often comes from experiments that follow a “shotgun” approach favoring coverage over accuracy. There are false positives and missing interactions and lots of interactions that occur under under experimental conditions but not in a cell. These limitations are all of particular concern for less-studied proteins with uncorroborated interactions, bacterial ABC proteins and their neighbors fall in this group. One of the main conceits of this project was to create multi-species interactomes to get a more reliable look at the interactions of ABC transporters and hopefully to find promising regulatory proteins.
Assayed protein interactions are supposed to mimic the interactions inside a cell so the data is specific to each species. However, because the ABC metal ion transport system is well conserved in most bacteria and any one species is likely to have incomplete or unreliable data, we set out to match homologous proteins between species to cross-reference interactions. This proved easier said than done as naming conventions between species were rarely descriptive and had little relation to each other. Matching homologous proteins using their names and meta data had little success. Querying other databases, like UniProt, ran into the same problems of incomplete or incompatible names and annotations when comparing between species. The interactomes we were able to generate had poor homology coverage which made it infeasible to rank interactions algorithmically and searching them by hand didn’t produce any obvious promising results.
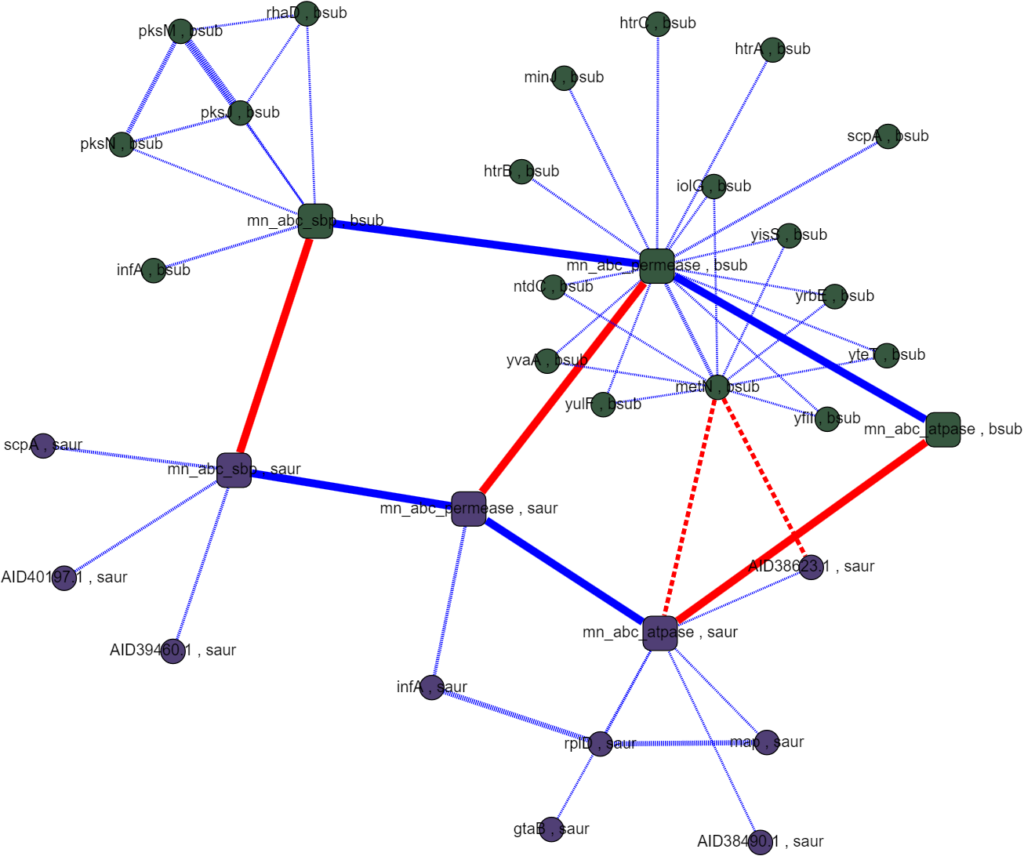
While this foray into multi-species interactomes didn’t yield compelling results for our research question, it was an exciting project with many takeaways for further research. Establishing homology between species will require more consistently rich data, like sequence data, and would likely benefit from the use of multiple methods including sequence alignments, ontology data, and annotated meta data. Also, while human legible models are very important for sanity checks and meaningfully interpreting the interactome with respect to a given research question, it can be a bit of a liability. In the course of this project the data and analyses included in the interactome were limited by what could be represented in the visualization when they didn’t have to be. Separating what was running “under the hood” from what was visualized with Graphspace would have allowed more freedom to include rigorous analyses. Factoring these considerations into future work promises a very useful tool for estimating protein-protein regulatory interactions in the ABC transport system and beyond!
