9/8/17
We are starting off the year neck deep in our literature review! First, we’re looking at how some researchers built the GIANT network, a functional interaction network built around tissue specificity 1. This type of network would be very useful to us given that we’re looking at neuronal motility genes, which needs to be specific to neurons since a ton of other cells move around.
How to Build a Tissue Specific Functional Gene Interaction Network
-
Don’t build a tissue specific functional gene interaction network
First, they built a “gold standard” of gene pairs that most definitely not tissue specific by selecting specific biological process gene collections from the Gene Ontology Consortium (GO). Gene pairs that were co-annotated to be functionally related were placed as the positive examples. Gene pairs that were not co-annotated to be functionally related were placed in the not-functionally-related-bin, unless they were: (A) in two different GO groups that had a significant number of shared genes, or (B) if the two genes were in GO groups that had nothing to do with each other. These gene pairs were ignored. They found 604,038 functionally related genes and 12,425,713 unrelated pairs.
-
Ignore the 13 million pair list you just made
Next, they matched Human Protein Reference Database gene to tissue annotations to the BRENDA Tissue Ontology. Tissues with less than ten genes were deemed worthless. They then grabbed a separate list of genes that are expressed ubiquitously and removed them from their respective sets of newly categorized genes and placed them into a “ubiquitous” bucket. Thus, they made a set of gene sets of genes expressed exclusively in specific tissues (T) and a set of genes expressed ubiquitously (U).
-
Integrate the Two lists
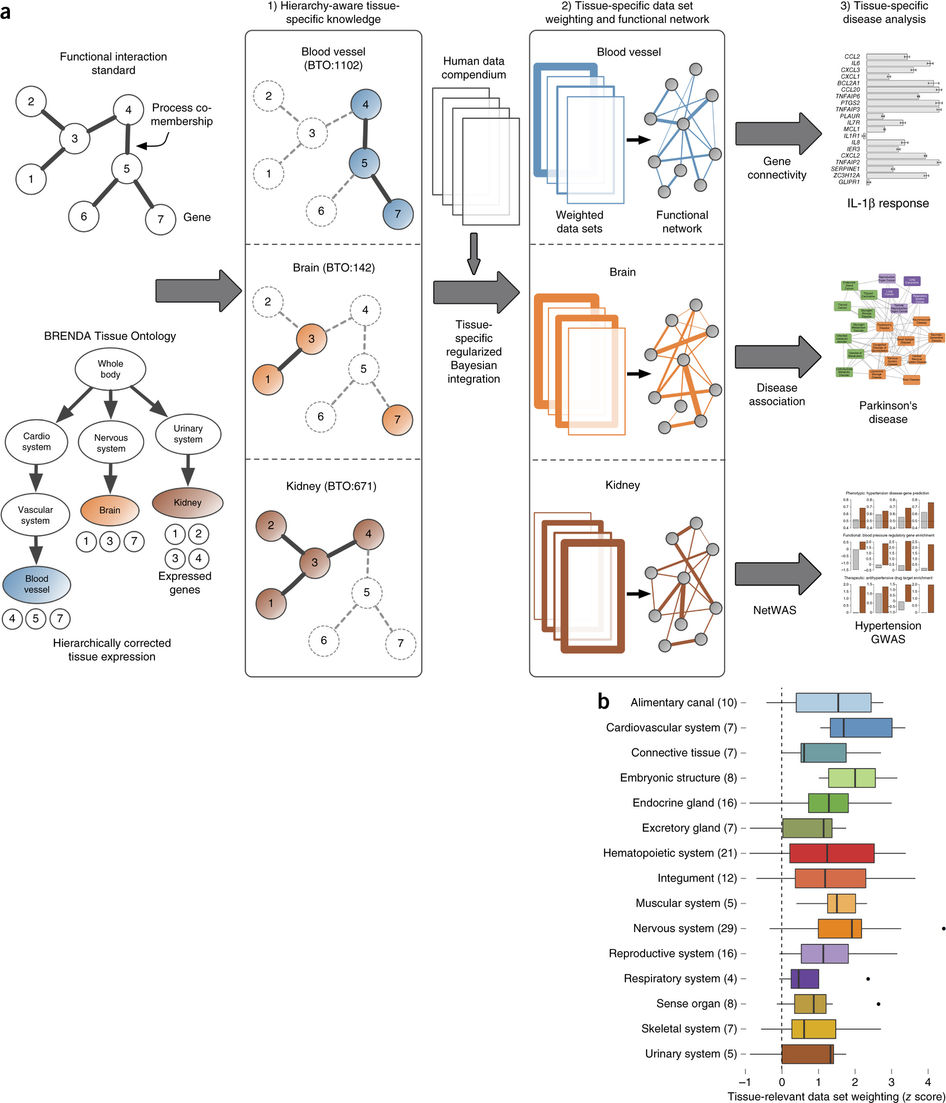
Greene et al, 2015
Going back to the tissue naive list of gene pairs, every gene was labeled depending on its tissue specificity. The gene pairs were then categorized according to each gene’s tissue specificity. The pairs with genes “specifically co-expressed in the tissue [T-T and T-U]” were marked as tissue specific, whereas the remaining gene pairs were marked as negative. The integrations were limited to 144 tissues with at least 10 positive tissue specific gene pairs.
4. Train a Bayesian Classifier for Each Tissue
They also trained a tissue naive classifier using the the original naive pair list. Since there was little independence, which is needed for Bayes classifiers, the dependency was calculated and accounted for. The classifiers then were able to make genome wide predictions about that specific tissue.
Hopefully I can learn more about the nuts and bolts of GIANT. These tools seem incredibly powerful, and might be used to find targets we might have never thought of. I will continue researching throughout the week.
- Greene CS, Krishnan A, Wong AK, Ricciotti E, Zelaya RA, Himmelstein DS, Zhang R, Hartmann BM, Zaslavsky E, Sealfon SC, Chasman DI, FitzGerald GA, Dolinski K, Grosser T, Troyanskaya OG. (2015). Understanding multicellular function and disease with human tissue-specific networks. Nature Genetics. 10.1038/ng.3259w. ↩︎
