This week, Anna drafted me to make a program to find the common genes in my schizophrenia dataset. It was a little tricky, given that all of the datasets from the different studies were organized differently, but it wasn’t anything I couldn’t handle.
Legend:
108 LOCI
Schizophrenia Working Group of the Psychiatric Genomics Consortium (2014). Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427.
SZGENE
Allen, N.C., Bagade, S., McQueen, M.B., Ioannidis, J.P.A., Kavvoura, F.K., Khoury, M.J., Tanzi, R.E., and Bertram, L. (2008). Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat Genet 40, 827–834.
HIPSC
Brennand, K.J., Simone, A., Jou, J., Gelboin-Burkhart, C., Tran, N., Sangar, S., Li, Y., Mu, Y., Chen, G., Yu, D., et al. (2011). Modelling schizophrenia using human induced pluripotent stem cells. Nature 473, 221–225.
DEADEXPRESSION
Fromer, M., Roussos, P., Sieberts, S.K., Johnson, J.S., Kavanagh, D.H., Perumal, T.M., Ruderfer, D.M., Oh, E.C., Topol, A., Shah, H.R., et al. (2016). Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci 19, 1442–1453.
CHROMOSOME CONFORMATION
Won, H., de la Torre-Ubieta, L., Stein, J.L., Parikshak, N.N., Huang, J., Opland, C.K., Gandal, M.J., Sutton, G.J., Hormozdiari, F., Lu, D., et al. (2016). Chromosome conformation elucidates regulatory relationships in developing human brain. Nature 538, 523–527.
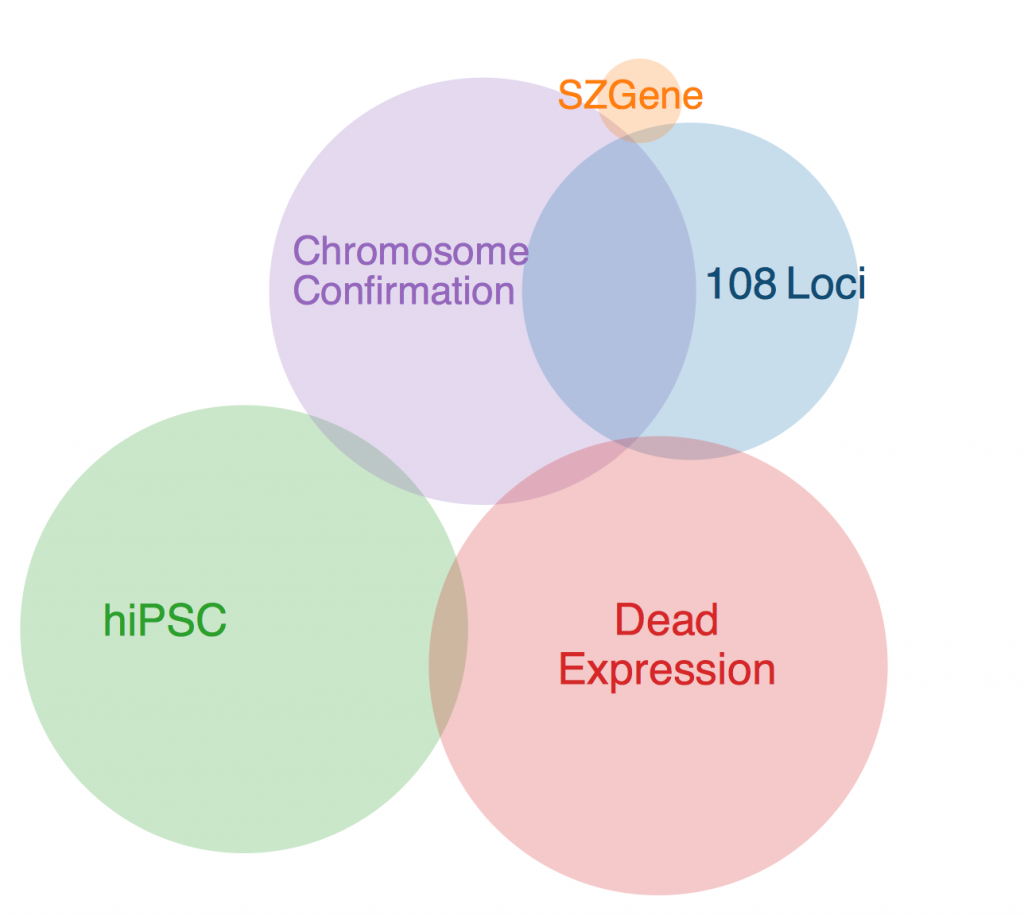
The first thing to note is that there was one gene that was in every data set except SZGene that couldn’t be included in this diagram because the Venn diagram program I found online deemed it geometrically impossible. This gene is TCF4, also known as immunoglobulin transcription factor 2. It is implicated in Pitt-Hopkins Syndrome and is involved in initiating neural differentiation.
An overlap between hiPSC, DeadExpression, and Chromosome Conformation is PRICKLE2, or Prickle Planar Cell Polarity Protein 2. It seems to be involved in the growth of post synaptic densities and neurite outgrowth.
Another pertinent overlap is EFHD1. EFHD1 was found in 108 Loci, hiPSC, and Chromosome Conformation. It seems to be calcium dependent and involved in apoptosis and neural differentiation, and it’s probably mitochondria dependent. However, its family is involved in cytoskeletal rearrangement.
One possibility for the dearth in overlap is simply the diversity in methods. Each collection got its genes from different areas, and it’s very likely that gene expression is totally different in all of these contexts. I’ll probably ask Anna more about this later. Until then, I will restart my Bayesian studies.
