We have passed the 90th consecutive night of Portland protests for Black Lives Matter. That’s three full months. That’s a whole summer.
There have been over 25,000 COVID-19 cases and 438 deaths in Oregon. The number of infected individuals total the entire population of Forest Grove, Happy Valley, or Wilsonville.
And yet, somehow, the students and staff who did research with me did phenomenally. Let me be clear: our success as a research group was driven by the collective effort of everyone involved.
What’s Published
We have had two papers and three posters accepted to the ACM Conference on Bioinformatics, Computational Biology, and Health Informatics (ACM-BCB).
- Tobias Rubel (Reed Spring/Fall ’19), now a post-baccalaureate working with me after graduation, will present our work on Augmenting Signaling Pathway Reconstructions.
- Ananthan Nambiar (Reed ’19), now a graduate student at UIUC, will present our work with computational linguist and Reed professor Mark Hopkins and UIUC co-authors Sergei Maslov, Maeve Heflin, and Simon Liu on Transformer Neural Networks for Protein Family Prediction Tasks.
- Gabe Preising (Reed ’20), now a post-baccalaureate working with me and Suzy Renn in the Biology Department, will present his undergraduate thesis work on building a Cichlid interactome and analyzing fish brain gene expression data to better understand maternal mouthbrooding.
- Frank Zhuang (rising Biology junior) will present his work on dissecting suffix/prefix variation in the Retinoic Acid Response Element (RARE) motif in zebrafish transcriptomes. This work built upon Tayla’s previous summer research.
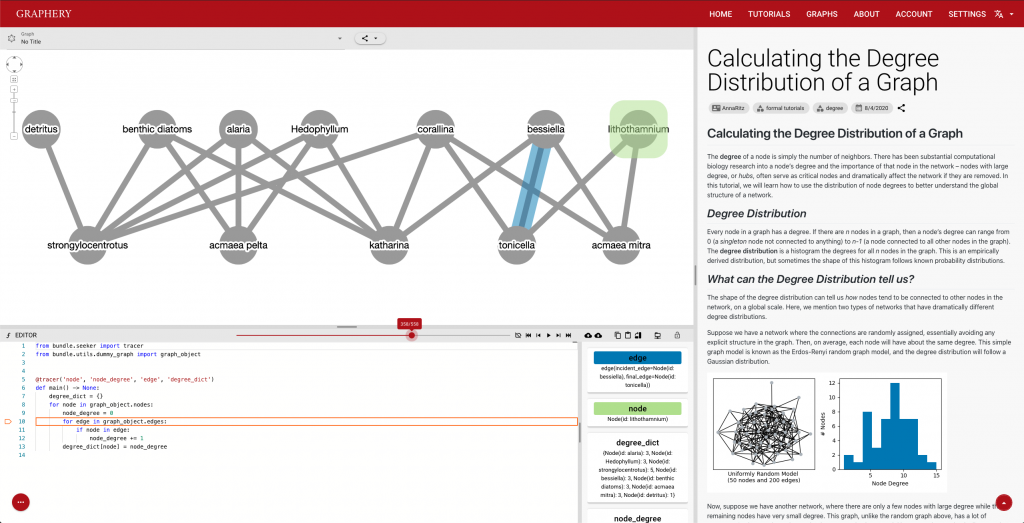
- Larry Zeng (rising Computer Science sophomore) built Graphery, a web-based graph tutorial platform for biologists to learn graph algorithms in the context of biological networks.
In addition to the ACM-BCB submissions, my collaborator Derek Applewhite’s group published work in Molecular Biology of the Cell (MBoC) on the RN-tre protein and its role in regulating a molecular motor, non-muscle myosin II, in Drosophila cells.
What’s Out
But wait, there’s more!
- Alex Richter (rising Math/CS junior) has re-implemented a hypergraph connectivity algorithm and it is currently being integrated into the ReactomeFIApp Cytoscape Plugin, which allows users to navigate Reactome using the Cytoscape graph visualization software. We have worked with collaborators Guanming Wu and Liam Beckman at Oregon Health & Sciences University on this integration effort.
- Jiarong Li (rising Math/CS senior) undertook a large cloud computing benchmark project, evaluating five platforms across about a dozen criteria for three application-specific tasks. Her final report will be shared with Reed’s computing technology staff as a recommendation for cloud computing options for faculty and students.
- Darsh Mandera, a rising sophomore at Jesuit High School, has posted a preprint on his project to apply machine learning techniques to predict drugs for cancer patients based on their genomic profile. Darsh has worked on this since middle school, and the project is close to his heart.
- My collaborator George Thomas at OHSU and colleagues have a preprint that identifies and evaluates drug combination therapies for kidney cancer. This paper is a massive undertaking by both cancer and computational biologists, and I was lucky to contribute a small part to this effort.
What’s Coming Down the Pipeline
Sometimes the most exciting projects are the ones that are just beginning.
- Aryeh Stahl (rising Math/CS senior) investigated pathway reconstruction algorithms when faced with experimental data from cancer patients. This changes the problem slightly; now, we wish to reconstruct pathways that have been dysregulated due to disease. His work on both real and simulated data may have uncovered a new way of thinking about pathway reconstruction in disease.
- Maham Zia (Reed ’20) was a post-baccalaureate researcher this summer before moving to join a lab at the University of Minnesota. Her work to automate cell fluorescence calculations in microscopy images is a promising start to automate common measurements related to cell imaging.
- I want to welcome postdoctoral researcher Pramesh Singh, who began work this week in my group. Pramesh is a physicist by training who has done work on complex networks, including biological networks – I am looking forward to working with him on new ideas.
More than Just Products
This summer was much, much more than just the computational biology work we produced. We were able to do some socially-distanced croquet to see each other face to face, which was definitely one of my summer highlights.

Finally, I want to thank the students and staff who have been actively and continually protesting for Black lives. You are an inspiration.

