My name is Tayla, and I’m a rising junior Biology major working on a research project co-advised by Anna Ritz and Kara Cerveny this summer. Overall, my project is trying to understand a vitamin A-dependent biological signaling pathway that is part of the process of stem cells differentiating into neurons.
We’re interested in this process because previous studies have shown that vitamin A is essential to proper embryonic eye development because it alters gene expression at the transcription level via specialized receptor proteins. Understanding this developmental process will provide insight into the complex differentiation process and identifying the involved genes in silico may open avenues of inquiry for in vivo studies. We hope to search for the genes that are affected by this pathway through sequence analysis and analyze how those genes might fit into this regulatory network.
But first, some background: the pathway that I’m looking at is the retinoic acid pathway which is especially significant in the retina of developing zebrafish embryos. Retinoic acid is an active metabolite of vitamin A that allows proteins to bind to DNA and alter the transcription of certain genes (Figure 1). These proteins, called retinoic acid receptors, alter in the presence of retinoic acid to bind to very specific DNA sequences called retinoic acid response elements (RAREs).
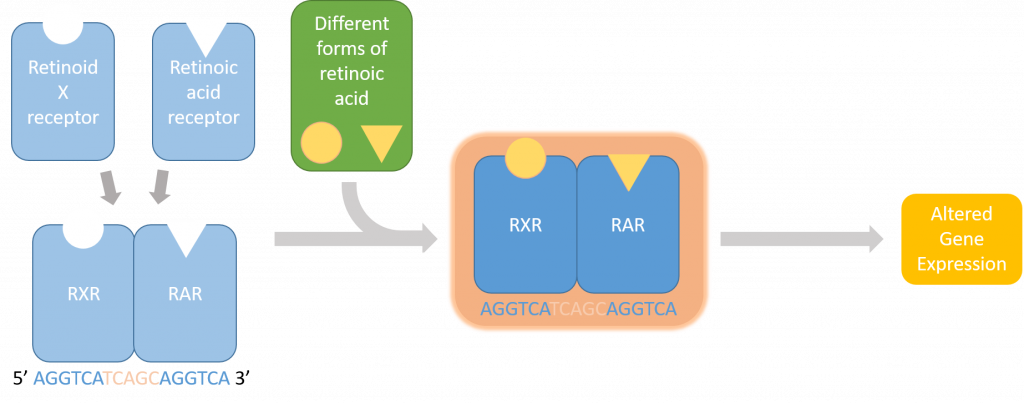
One of my goals for this project is to find zebrafish genes that are responsive to retinoic acid influxes. To do this, I have to scan through parts of the genome and look for a tandem repeat of a six base pair motif. Retinoic acid receptors bind to these RAREs within the sequence upstream of the affected gene. I can build a program that takes these upstream regions of zebrafish genes, finds this repeated motif, and tells me all the genes that were found.

While it sounds pretty simple, there are actually a lot of moving parts. First I have to read in a big file of sequence and identifying information, and preferably do it quickly. Then I have to find a six base pair motif repeated 1-5 base pairs downstream and score it according to what’s allowed by its documented variation (Figure 2). Finally I have to return the gene IDs of genes containing the repeat. All of this is run on 65,171 annotated zebrafish transcripts’ upstream regions.
Luckily, at this point in my project (about 6 weeks in), I’ve written a program that will do this in about half an hour. Now comes the interpretation: finding out where and at what stage the genes I identified with my program are expressed in zebrafish. Hopefully we’ll find some genes that we expect to be regulated by retinoic acid in the final set of candidates to validate our method. The most exciting prospect is perhaps finding novel genes regulated by this pathway, or better yet a confirmation that the genes we’re testing in the lab as direct targets of retinoic acid exhibit the canonical response site.
Sources:
Al Tanoury Z, Piskunov A, Rochette-Egly C. Vitamin A and retinoid signaling: genomic and nongenomic effects. J Lipid Res. 2013;54(7):1761-1775. doi:10.1194/jlr.R030833
Cunningham TJ, Duester G. Mechanisms of retinoic acid signalling and its roles in organ and limb development. Nat Rev Mol Cell Biol. 2015;16(2):110-123. doi:10.1038/nrm3932
Lalevée S, Anno YN, Chatagnon A, et al. Genome-wide in Silico Identification of New Conserved and Functional Retinoic Acid Receptor Response Elements (Direct Repeats Separated by 5 bp). J Biol Chem. 2011;286(38):33322-33334. doi:10.1074/jbc.M111.263681
Predki PF, Zamble D, Sarkar B, Giguère V. Ordered binding of retinoic acid and retinoid-X receptors to asymmetric response elements involves determinants adjacent to the DNA-binding domain. Mol Endocrinol. 1994;8(1):31-39. doi:10.1210/mend.8.1.8152429
