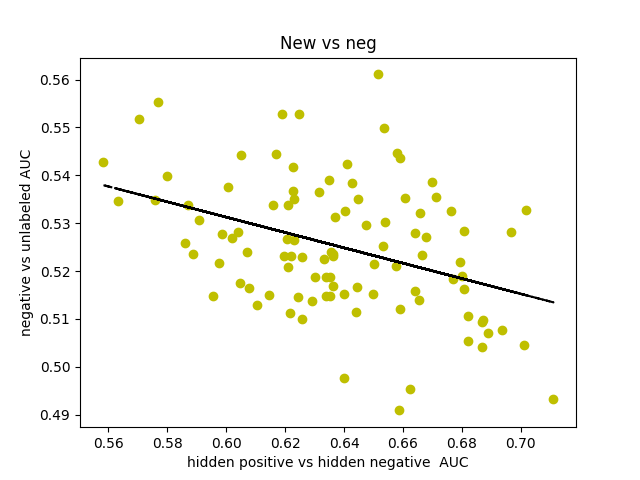
I was curious about what our AUC would look like if instead of comparing hidden positives to unlabeled, we compared hidden positives to hidden negatives, wondering if that would get us a more valid representation of the network’s ability to predict genes associated with schizophrenia. What if the unlabeled genes ranked above the hidden positives were simply actual centers of genetic perturbations in schizophrenia?
Oddly enough, the scores actually decreased from 0.65-0.70 to 0.60-0.65. I found that the AUCs for comparing hidden negatives to unlabeled nodes hovered around 0.53, meaning that the hidden negatives were ranked mostly in the top half of the list.
This somewhat makes sense though. The basis for the negative set is that they are genes in non-neurological diseases, but that doesn’t necessarily exclude them from being included in a more subtle, polygenic disorder that’s dependent on additive effects of hundreds of genetic perturbations. It also makes sense that genes involved in other diseases might have a slightly higher probability of being associated with some other disease.
I decided to make a new negative set. I started by gathering every single node in the 0.150 network. There is a resource called SZGR 2.0 (https://bioinfo.uth.edu/SZGR/) that collects all sorts of evidence for genes being associated with schizophrenia. Using this list, I excluded any gene that had any evidence for being associated with schizophrenia. I then took differential gene expression data from autism spectrum disorder, bipolar disorder, and major depressive disorder as well as schizophrenia (http://science.sciencemag.org/content/359/6376/693). If a gene was not differentially expressed in any of these diseases (FDR>0.5 for schizophrenia, FDR>0.2 for others) and was in the other list, I added that gene to my list of negatives, which was 1561 genes long. I found that this list, when hidden negatives were compared to unlabeled genes, had an AUC of about 0.44, which means that our negatives are finally oriented around schizophrenia rather than non-neurological diseases. I also found that the AUC for hidden positives vs hidden negatives spiked up to 0.75, meaning that our program is legitimately good at predicting genes associated with schizophrenia.
I found no significant performance differences from factoring in evidence levels.
