The week before break, at the group meeting, we discussed the gene lists Miriam and I created. Because of the lack of overlap on my part, my next job to do is to modify my program to account for possible different gene names. Anna sent me a giant text file for it, and I will get it done by the Friday meeting.
I did not spend all of break dormant, and I learned more about bayesian statistics in addition to a brief overview by one of Anna’s post-docs. I think one of the difficulties of knowing how Bayes’ theorem works is the fact that it’s just so ingrained into our normal thought. Given B, what is the probability of A? It’s the probability of B given A times the probability of A divided by the probability of B. The first thing to note is the B denominating the whole equation. The B is accounting for the probability warping from the context of the problem. The second factor is probability of B given A. This represents the relationship we already know. This is multiplied by the probability of A. Therefore, the numerator represents the total probability of B happening because of A, which can also be described as the total probability that the specified relationship happens. By accounting for the probability warping in the denominator, we get the actual probability of A given B.
Bayesian probability is the core of the functional interaction network and the integrated network we will make. I can already kind of see how gene interaction probabilities could be derived from this given interaction data.
However, the mutual exclusivity clause in the theorem might be tricky. I’ll have to closely look at the supplemental data to see how the functional interactive builders accounted for this.
Week 6
Currently on break. Will resume blog posts next week.
Week 5
This week, Anna drafted me to make a program to find the common genes in my schizophrenia dataset. It was a little tricky, given that all of the datasets from the different studies were organized differently, but it wasn’t anything I couldn’t handle.
Legend:
108 LOCI
Schizophrenia Working Group of the Psychiatric Genomics Consortium (2014). Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427.
SZGENE
Allen, N.C., Bagade, S., McQueen, M.B., Ioannidis, J.P.A., Kavvoura, F.K., Khoury, M.J., Tanzi, R.E., and Bertram, L. (2008). Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat Genet 40, 827–834.
HIPSC
Brennand, K.J., Simone, A., Jou, J., Gelboin-Burkhart, C., Tran, N., Sangar, S., Li, Y., Mu, Y., Chen, G., Yu, D., et al. (2011). Modelling schizophrenia using human induced pluripotent stem cells. Nature 473, 221–225.
DEADEXPRESSION
Fromer, M., Roussos, P., Sieberts, S.K., Johnson, J.S., Kavanagh, D.H., Perumal, T.M., Ruderfer, D.M., Oh, E.C., Topol, A., Shah, H.R., et al. (2016). Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci 19, 1442–1453.
CHROMOSOME CONFORMATION
Won, H., de la Torre-Ubieta, L., Stein, J.L., Parikshak, N.N., Huang, J., Opland, C.K., Gandal, M.J., Sutton, G.J., Hormozdiari, F., Lu, D., et al. (2016). Chromosome conformation elucidates regulatory relationships in developing human brain. Nature 538, 523–527.
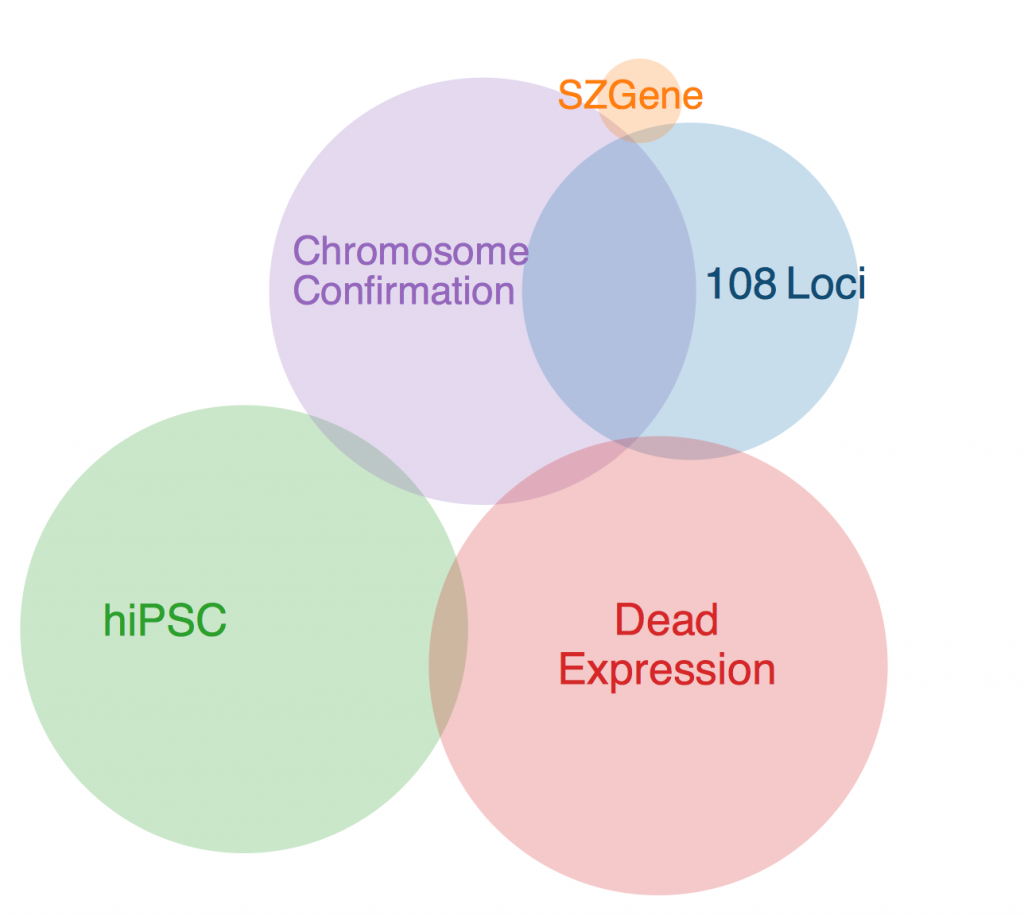
The first thing to note is that there was one gene that was in every data set except SZGene that couldn’t be included in this diagram because the Venn diagram program I found online deemed it geometrically impossible. This gene is TCF4, also known as immunoglobulin transcription factor 2. It is implicated in Pitt-Hopkins Syndrome and is involved in initiating neural differentiation.
An overlap between hiPSC, DeadExpression, and Chromosome Conformation is PRICKLE2, or Prickle Planar Cell Polarity Protein 2. It seems to be involved in the growth of post synaptic densities and neurite outgrowth.
Another pertinent overlap is EFHD1. EFHD1 was found in 108 Loci, hiPSC, and Chromosome Conformation. It seems to be calcium dependent and involved in apoptosis and neural differentiation, and it’s probably mitochondria dependent. However, its family is involved in cytoskeletal rearrangement.
One possibility for the dearth in overlap is simply the diversity in methods. Each collection got its genes from different areas, and it’s very likely that gene expression is totally different in all of these contexts. I’ll probably ask Anna more about this later. Until then, I will restart my Bayesian studies.
Weeks 4 and 5: Building a Gold Standard List
Over the past couple of weeks, Alex and I have been given the task of building our project’s gold standard list. This list will be comprised of genes known to be associated with Schizophrenia and genes known to be associated with cell motility; Alex was given the Schizophrenia half to research and I was given the cell motility half. Overwhelmed by the sheer volume of genes and pathways associated with cell motility, I began by looking up genes known to be associated with both Schizophrenia and cell motility. In our meeting, we briefly discussed how this might serve us later as a known positive we could use to test our algorithm.
I read through tons of articles and papers, many of which I found through references in previous papers we have read. In reading these papers, I was hunting for genes, pathways, and resources that were studied and employed by the researchers and found several that will be useful to us. The CAM pathway and associated genes appeared multiple times, and the resource KEGG was used by the majority of the researchers. My task over the past week has been to download data from KEGG, continue accumulating genes for our gold standard list, and begin adding “confidence details” to the genes – this is an important weight that will be added to the genes in our list and will be a measure of how “confident” we are that the gene is truly associated with either Schizophrenia or cell motility.
Week 4
I am currently in the process of figuring out which genes are common to each of the data sets. This will involve building a program to find the common genes. I have a good idea of how to do it; it’s a relatively simple problem. The only hangups will probably be translating all the datasets into one universal comparable format, but given enough time to iron out the kinks in all 5 data sets, I think I can manage easily. Eventually, I’ll make a neat Venn diagram illustrating what genes are common to what datasets.
I’m also learning more and more about bayesian probability. This is the basis of the functional interaction network we will eventually use. Anna’s postdoc Ibrahim explained the basics to me. This is a new concept to me, so this week, I’ll also learn more about that as well.
Week 3
This week, we searched for genes related to both schizophrenia and cell motility in order to build out gold standard of genes upon which to base our program off of.
My job was to find collections of genes related to Schizophrenia (SZ), while Miriam searched for cell motility genes. Our program is planned to have the capability to weight evidence for the strength of the data, so even uncertain genes will be really helpful.
The first set of associated genetic information is from the Schizophrenia Working Group of the Psychiatric Genomics Consortium. In a Genome Wide Association Study (GWAS) from 36,989 cases, they identified 108 loci that contained SNPs significantly more likely to be present in people with SZ. The threshold for significance is a p value of less than 5×10^-8, making this study one of the strongest sets of SZ gene data. However, while most of these SNPs are located near protein coding genes, all but 10 are located within non-coding regions. A regulatory region being near a protein coding gene does not necessarily mean that the gene is actually regulated by that region. Instead, the region might regulate some other gene in some other area. However, regionality with a regulator does imply a higher probability of being regulated by the regulator, so this locational information might be useful in the weaker evidence standards.
http://www.nature.com/nature/journal/v511/n7510/abs/nature13595.html?lang=en
The next set of data is from the Schizophrenia Research Forum on SZGene.org. It contains every genetic association study paper for SZ genes that’s available. It’s a nice collection, but none of the genes reached the GWAS p value threshold of 10^-8. This evidence will have to be weighted proportional to the strength of the studies themselves.
Another set of data comes from an expression study on human induced pluripotent stem cells. The cells were differentiated to neurons and underwent qPCR. This data, however, is in vitro, whereas the developmental aspect of SZ requires a degree of specificity and communication. https://www.nature.com/nature/journal/v473/n7346/full/nature09915.html
There is another gene expression dataset, but this time, it’s from dead people. While the region of the brain is much more specific (prefrontal cortex), this expression data comes long after development. However, they did find 1 gene, inhibited it in a pluripotent stem cell, and found abnormal cell motility. Therefore, this data may be useful to our study. http://www.nature.com/neuro/journal/v19/n11/full/nn.4399.html
The last genetic data set covers the conformational structure of the chromatin in the nucleus. Brain specific intrachromatin contacts can upregulate protein coding genes promoted by the contact. Thus, they established a strong correlation between gene expression and chromatin contact using 3 neonatal brain slices from separate subjects. They were able to locate the points of contact for the remaining 98 loci in the initial gene, giving a more broad picture of genetic interaction. http://www.nature.com/nature/journal/v538/n7626/full/nature19847.html
We’ll probably use all of these data sets. The next question is how.
Week 3
No blog post this week! We didn’t have our regular meeting due to the PacNow QB Meeting hosted by Anna and Derek. This week Alex and I are presenting an overview of our project as well as the papers we’ve read so far.
Week 2
This week we looked at a paper describing the relationship between cellular focal adhesions, cell motility, and schizophrenia. http://www.sciencedirect.com/science/article/pii/S0006322313000917
Previous gene expression assays from olfactory mucosa neurons, which are excellent for disease modeling because of their multipotentcy, from patients with schizophrenia found differential expression in focal adhesion related genes, such as integrin genes (http://dmm.biologists.org/content/3/11-12/785.long). Differential expression was also found in the Focal Adhesion Kinase (FAK) pathway, which is central to the construction and maintenance of Focal Adhesions. Focal Adhesions are necessary for binding the cell to the extracellular matrix, which keeps it lodged in one place. Focal Adhesions are also important for cell motility insofar as cells need some sort of friction to move around. The fatty acid lipid membrane is very flexible and slippery due to its hydrophobicity; it needs the focal adhesions to provide an equal and opposite force for cellular motion. Imagine you’re on an ice rink, but your shoes can penetrate the ice and grip it so that you can walk.
However, these researchers found an increased level of cell motility in neurons of schizophrenic patients. Schizophrenic cells are faster and travel farther than control cells. Schizophrenic cells were also found to be less adherent, with less cells binding to plates when creating cell cultures. They found a reduced level of phosphylated FAK (pFAK) in patient cells but found that inhibiting FAK returned cell motility to control levels. They also found that inhibiting α3β1 integrins and α8β1 integrins also returned motility to normal. This implies that instead of the reduced level of FAK being the cause of the increased motility, the cause is instead a malfunction in the FAK pathway itself, of which α3β1 integrin and α8β1 integrin are a part of. They also found that patient focal adhesions were smaller, less common, and disassembled faster.
Unfortunately, there are an enormous amount of genes involved in cell motility and the FAK pathway. Any one of them could be affecting FAK, integrins, and cell motility at large. Hopefully our functional network programs will help elucidate some suspects.
This coming week, my primary goal will be identifying genes and pathways prominent in Schizophrenia pathology, while Miriam will be finding ones associated with cell motility
Week 2: FAK and Schizophrenia
This week we took a closer look at a paper that investigated cell adhesion, cell motility, and focal adhesion dynamics in Schizophrenia patients.
These migration functions are regulated by focal adhesion kinase (FAK) proteins. The focal adhesion kinase signaling pathway involves the expression of integrin genes; integrins are proteins that detect cell adhesion as well as attach the cell to the extracellular matrix during cell migration. A previous study showed that the expression of two integrin genes (ITGA8, ITGA3) was altered in schizophrenia-derived cells.
The study was conducted on olfactory neurosphere-derived cells (accessible via biopsy of the olfactory mucosa) from 9 healthy male subjects and 9 male schizophrenia patients using two different assays. The assays involved seeding the cells on fibronectin-coated plates or chambers, allowing them to attach for 4 hours, washing away non-adherent cells, and allowing remaining cells to migrate for different periods of time. These experiments were repeated with the presence of FAK phosphorylation inhibitors, and then again while blocking antibodies to two different types of integrins. Several pieces of data were analyzed, including levels of pFAK, migration distance, speed, and size and number of focal adhesions present in the cells.
The results of this study demonstrated several important disparities that we are paying close attention to. While there was no difference in levels of FAK between patients and control subjects, patients had significantly lower levels of phosphorylated FAK (pFAK). In addition, patient cells had fewer adhesions, were less adherent, and were more motile than control cells, with a higher percentage of patient cells migrating further and with greater speed. When pFAK was inhibited, and antibodies to two different types of integrins were blocked (three separate experiments), patient cell motility was reduced to control levels but control levels were not changed. This last result led us to conclude that the phosphorylation of FAK does not work like an on/off switch; rather, phosphorylation of FAK alters its behavior.
This paper gave us lots of food for thought; it provided us with a starting place to form our list of schizophrenia genes to investigate. Next week we will be investigating cell motility further by finding cell motility gene databases and papers studying the link between cell motility and schizophrenia.
Week 1: A Little Context
Week 1! We’re starting off the year, and our research, with some literature review to provide ourselves with a little context. Our proposed research plan mainly draws on two papers that laid out the methodological groundwork for us.
The first paper provides pertinent results and insights into the connection between cell motility and schizophrenia. The researchers concluded that cells in patients with schizophrenia are less adhesive and more motile than the cells of healthy control subjects, which was shown to improve with the inhibition of the focal adhesion kinase (FAK) protein. The results of this paper showed that there is a correlation between the altered motility of patient cells and dysregulated gene expression in the FAK signaling pathway within these cells. This paper provided us with the informational basis necessary to choose what kinds of genes we will shine our focus on – schizophrenia and migration association genes.
The second paper essentially laid out the framework for our methodology. In this paper, the researchers used a machine-learning algorithm to predict genes associated with autism spectrum disorder (ASD). They then validated these predicted genes experimentally using an independent-case sequencing study and were further able to demonstrate that this large set of ASD genes played roles in key pathways and brain development.
Pulling from both these papers, we plan to develop a network diffusion algorithm to identify candidate schizophrenia and migration associated genes. We are going to computationally build a list of predicted genes and (hopefully) validate them experimentally. However, before we do that, we need to delve into the nitty gritty of how the autism paper’s machine-learning algorithm was created.
In the paper, it outlines how the approach was based upon a human brain-specific gene functional-interaction network nicknamed GIANT (Genome-scale Integrated Analysis of gene Networks in Tissues). We began with a few initial questions: How is a functional-interaction standard set up? How was GIANT constructed? These questions will no doubt open up a whole new corridor of doors to explore; over the next couple of weeks, our goal is to investigate these questions through literature review and learn more about how we can utilize these tools in the coming year.
